 |
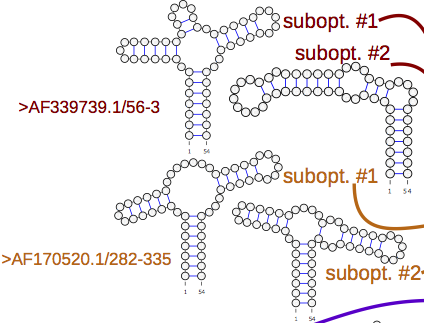
aliFreeFold & aliFreeFoldMulti
A program to predict the conserved secondary structures of homologous ribonucleic acid (RNA) sequences using an alignment-free approach. Stand-alone application + Webserver
|
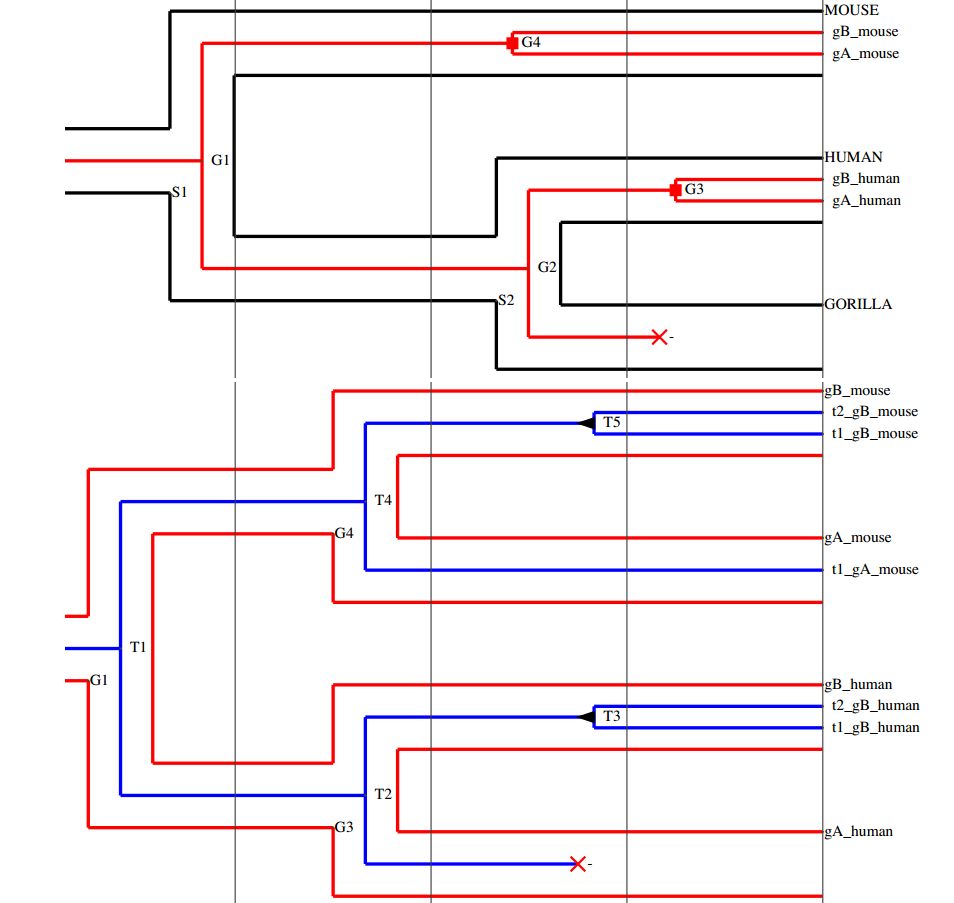 |
DoubleRecViz
A program for visualizing and editing double reconciliations between phylogenetic trees at three levels: transcript, gene and species. Stand-alone application + Webserver
|
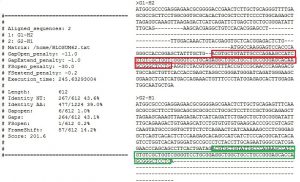 |
FsePSA
A program to compute pairwise alignments between coding DNA sequences while accounting for the length of frameshift translations. Stand-alone application + Webserver
|
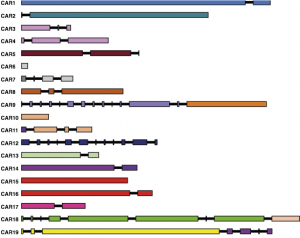 |
ProCARs
A program to reconstruct Ancestral gene orders as CARs (Contiguous Ancestral Regions) with a progressive homology-based method. Stand-alone application
|
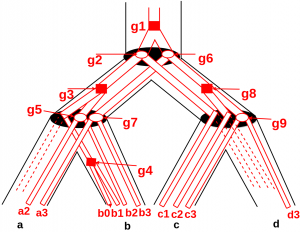 |
Protein2GeneTree
A program to compute a genetree minimizing the sum of reconciliation costs with a protein tree and a species tree. Stand-alone application + Webserver
|
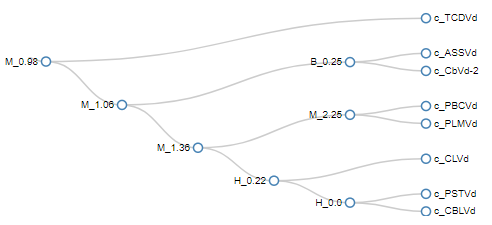 |
RNAFamViz
A tool for visualizing the evolution of RNA secondary structure motifs in a RNA family. Webserver
|
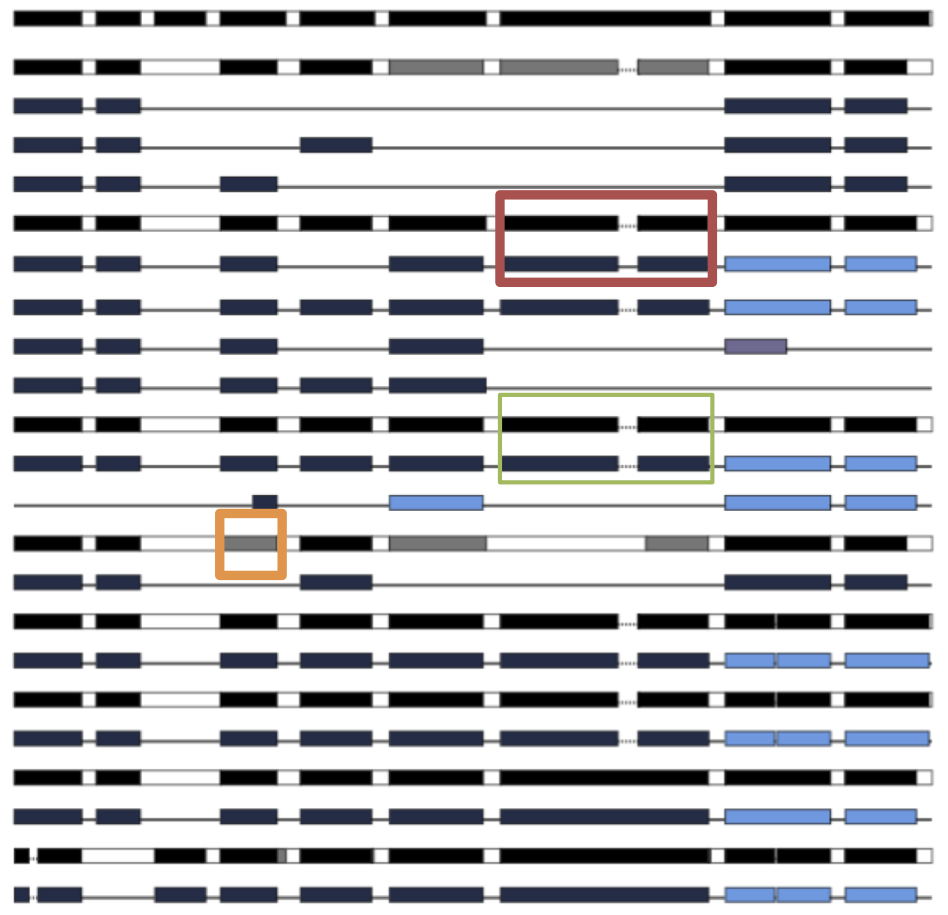 |
SpliceFamViz
A program to compute and visualize the multiple spliced alignments of transcript and gene sequences from a gene family. Webserver
|
 |
SpliceAlnViz
An interactive visualization tool for multiple spliced alignment of genes and alternative transcripts of a gene family. Stand-alone application + Webserver
|
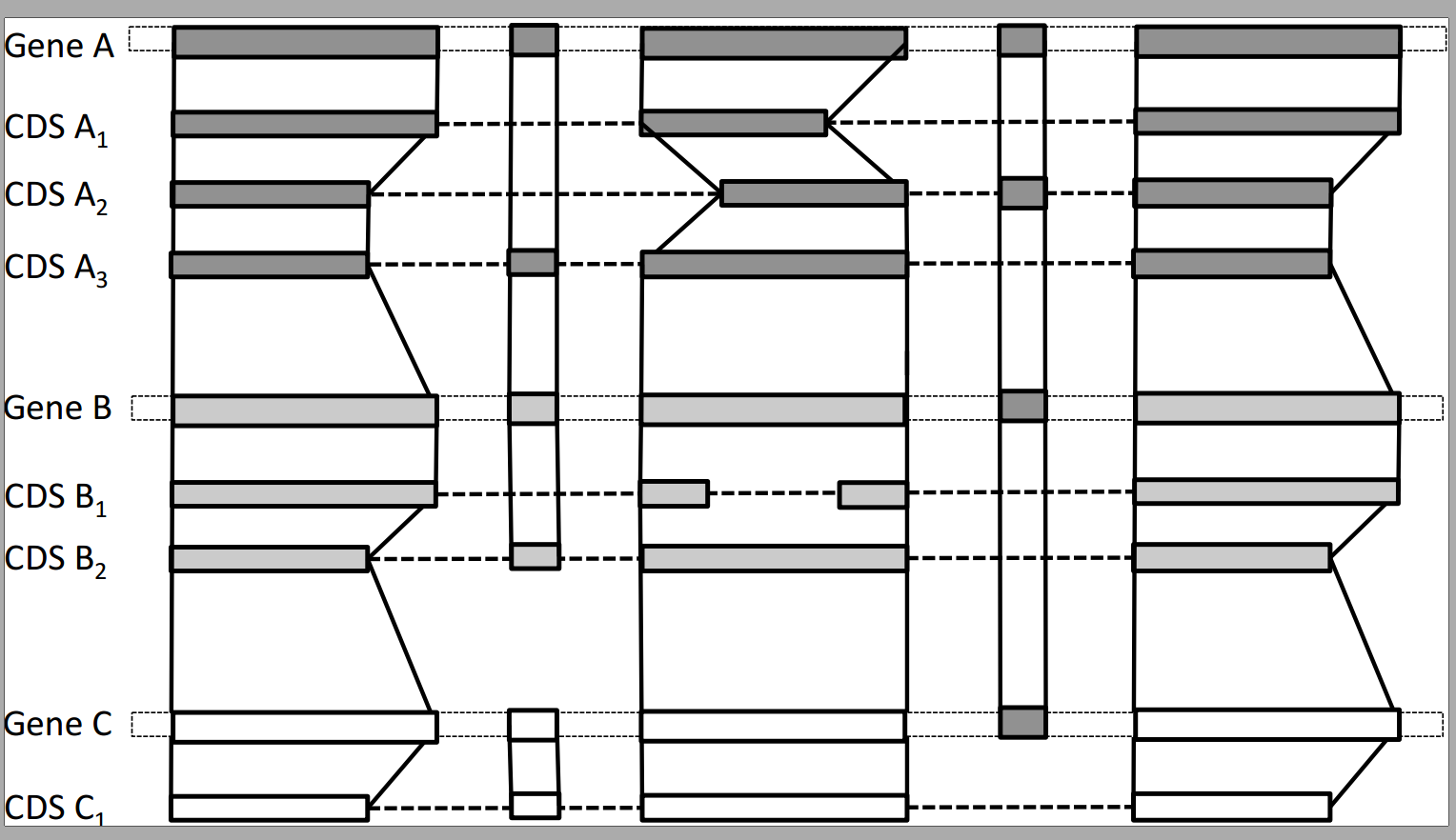 |
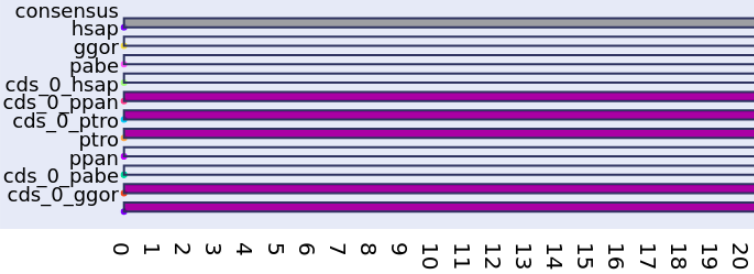
SpliceFamAlign & SpliceFamAlignMulti
A tool for computing the alignment of a spliced CDS against a gene sequence while accounting for the splicing structures of the input sequences, and then the inference of transcript splicing orthology groups in a gene family based on spliced alignments. Stand-alone application + Webserver
|
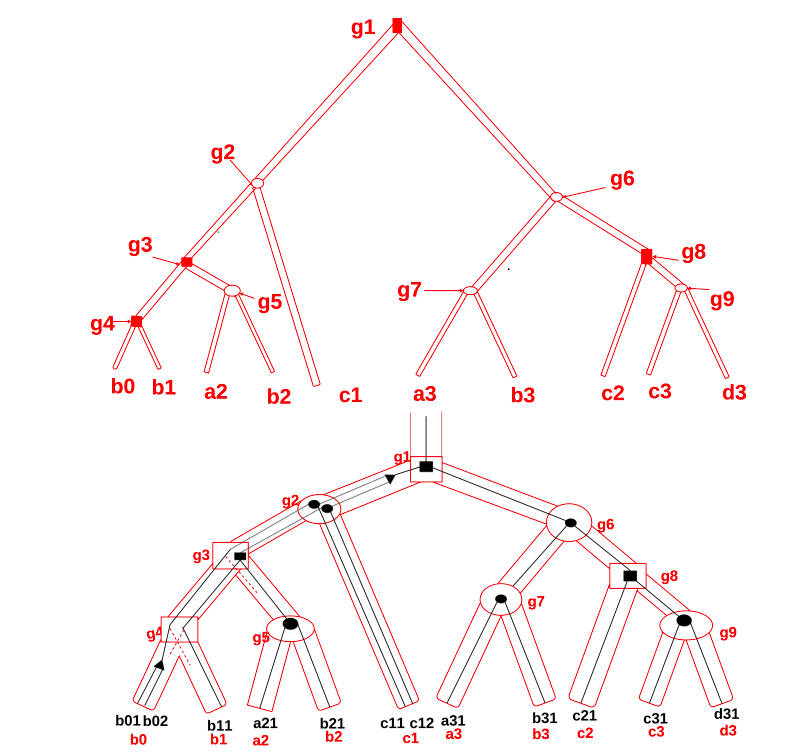 |
SuperProteinTree
A tool that computes a super protein tree given a set of protein clusters. It also reconstructs a gene tree using the double reconciliation with a protein tree and a species tree. Stand-alone application + Webserver
|
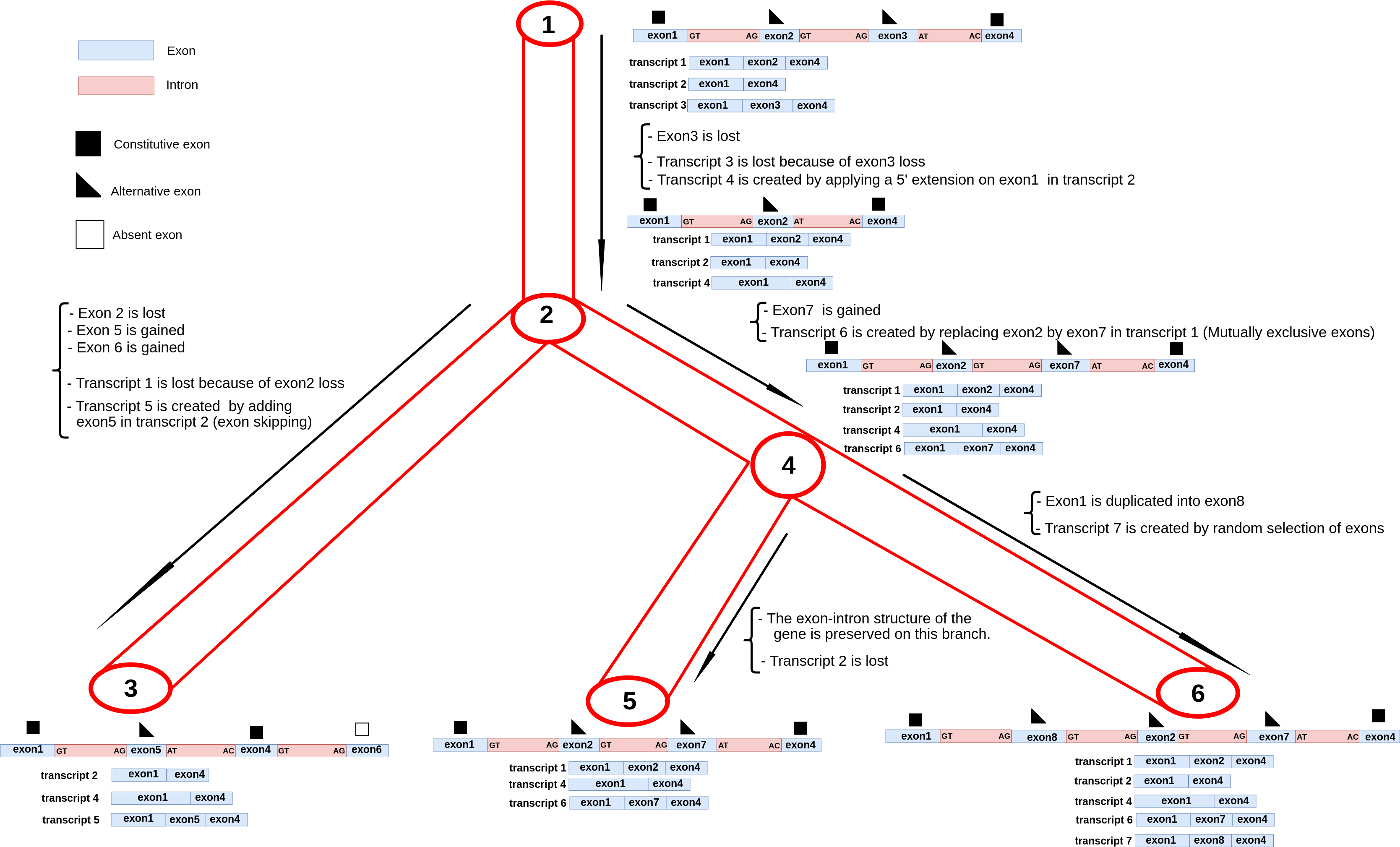 |
SimSpliceEvol
A new method for simulating the evolution of sets of alternative transcripts along the branches of an input gene tree. Stand-alone application + Webserver
|
